Plotting¶
This page describes in depth the general plotting capabilities of GCPy, including possible argument values for every plotting function.
For information about GCPy functions that are specific to the GEOS-Chem benchmark workflow, please see our Benchmarking chapter.
Six-panel comparison plots¶
The functions listed below generate six-panel plots comparing variables between two datasets:
Plotting function |
Located in GCPy module |
|---|---|
|
|
|
|
Both compare_single_level() and compare_zonal_mean()
generate a six panel plot for each variable passed. These plots can
either be saved to PDFs or generated sequentially for visualization in
the Matplotlib GUI using matplotlib.pyplot.show().
Each plot uses data passed from a reference (Ref) dataset
and a development (Dev) dataset. Both functions share
significant structural overlap both in output appearance and code
implementation.
You can import these routines into your code with these statements:
from gcpy.plot.compare_single_level import compare_single_level
from gcpy.plot.compare_zonal_mean import compare_zonal_mean
Each panel has a title describing the type of panel, a colorbar for
the values plotted in that panel, and the units of the data plotted in
that panel. The upper two panels of each plot show actual values from
the Ref (left) and Dev (right) datasets for a
given variable. The middle two panels show the difference
(Dev - Ref) between the values in the Dev
dataset and the values in the Ref dataset. The left middle
panel uses a full dynamic color map, while the right middle panel caps
the color map at the 5th and 95th percentiles. The bottom two panels
show the ratio (Dev/Ref) between the values in the Dev
dataset and the values in the Ref Dataset. The left bottom panel uses
a full dynamic color map, while the right bottom panel caps the color
map at 0.5 and 2.0.
Function compare_single_level¶
The compare_single_level function accepts takes the following
arguments:
def compare_single_level(
refdata,
refstr,
devdata,
devstr,
varlist=None,
ilev=0,
itime=0,
refmet=None,
devmet=None,
weightsdir='.',
pdfname="",
cmpres=None,
match_cbar=True,
normalize_by_area=False,
enforce_units=True,
convert_to_ugm3=False,
flip_ref=False,
flip_dev=False,
use_cmap_RdBu=False,
verbose=False,
log_color_scale=False,
extra_title_txt=None,
extent=None,
n_job=-1,
sigdiff_list=None,
second_ref=None,
second_dev=None,
spcdb_dir=os.path.dirname(__file__),
sg_ref_path='',
sg_dev_path='',
ll_plot_func='imshow',
**extra_plot_args
):
"""
Create single-level 3x2 comparison map plots for variables common
in two xarray Datasets. Optionally save to PDF.
Args:
refdata: xarray dataset
Dataset used as reference in comparison
refstr: str
String description for reference data to be used in plots
devdata: xarray dataset
Dataset used as development in comparison
devstr: str
String description for development data to be used in plots
Keyword Args (optional):
varlist: list of strings
List of xarray dataset variable names to make plots for
Default value: None (will compare all common variables)
ilev: integer
Dataset level dimension index using 0-based system.
Indexing is ambiguous when plotting differing vertical grids
Default value: 0
itime: integer
Dataset time dimension index using 0-based system
Default value: 0
refmet: xarray dataset
Dataset containing ref meteorology
Default value: None
devmet: xarray dataset
Dataset containing dev meteorology
Default value: None
weightsdir: str
Directory path for storing regridding weights
Default value: None (will create/store weights in
current directory)
pdfname: str
File path to save plots as PDF
Default value: Empty string (will not create PDF)
cmpres: str
String description of grid resolution at which
to compare datasets
Default value: None (will compare at highest resolution
of ref and dev)
match_cbar: bool
Set this flag to True if you wish to use the same colorbar
bounds for the Ref and Dev plots.
Default value: True
normalize_by_area: bool
Set this flag to True if you wish to normalize the Ref
and Dev raw data by grid area. Input ref and dev datasets
must include AREA variable in m2 if normalizing by area.
Default value: False
enforce_units: bool
Set this flag to True to force an error if Ref and Dev
variables have different units.
Default value: True
convert_to_ugm3: bool
Whether to convert data units to ug/m3 for plotting.
Default value: False
flip_ref: bool
Set this flag to True to flip the vertical dimension of
3D variables in the Ref dataset.
Default value: False
flip_dev: bool
Set this flag to True to flip the vertical dimension of
3D variables in the Dev dataset.
Default value: False
use_cmap_RdBu: bool
Set this flag to True to use a blue-white-red colormap
for plotting the raw data in both the Ref and Dev datasets.
Default value: False
verbose: bool
Set this flag to True to enable informative printout.
Default value: False
log_color_scale: bool
Set this flag to True to plot data (not diffs)
on a log color scale.
Default value: False
extra_title_txt: str
Specifies extra text (e.g. a date string such as "Jan2016")
for the top-of-plot title.
Default value: None
extent: list
Defines the extent of the region to be plotted in form
[minlon, maxlon, minlat, maxlat].
Default value plots extent of input grids.
Default value: [-1000, -1000, -1000, -1000]
n_job: int
Defines the number of simultaneous workers for parallel
plotting. Set to 1 to disable parallel plotting.
Value of -1 allows the application to decide.
Default value: -1
sigdiff_list: list of str
Returns a list of all quantities having significant
differences (where |max(fractional difference)| > 0.1).
Default value: None
second_ref: xarray Dataset
A dataset of the same model type / grid as refdata,
to be used in diff-of-diffs plotting.
Default value: None
second_dev: xarray Dataset
A dataset of the same model type / grid as devdata,
to be used in diff-of-diffs plotting.
Default value: None
spcdb_dir: str
Directory containing species_database.yml file.
Default value: Path of GCPy code repository
sg_ref_path: str
Path to NetCDF file containing stretched-grid info
(in attributes) for the ref dataset
Default value: '' (will not be read in)
sg_dev_path: str
Path to NetCDF file containing stretched-grid info
(in attributes) for the dev dataset
Default value: '' (will not be read in)
ll_plot_func: str
Function to use for lat/lon single level plotting with
possible values 'imshow' and 'pcolormesh'. imshow is much
faster but is slightly displaced when plotting from
dateline to dateline and/or pole to pole.
Default value: 'imshow'
extra_plot_args: various
Any extra keyword arguments are passed through the
plotting functions to be used in calls to pcolormesh() (CS)
or imshow() (Lat/Lon).
"""
and generates a comparison plot such as:
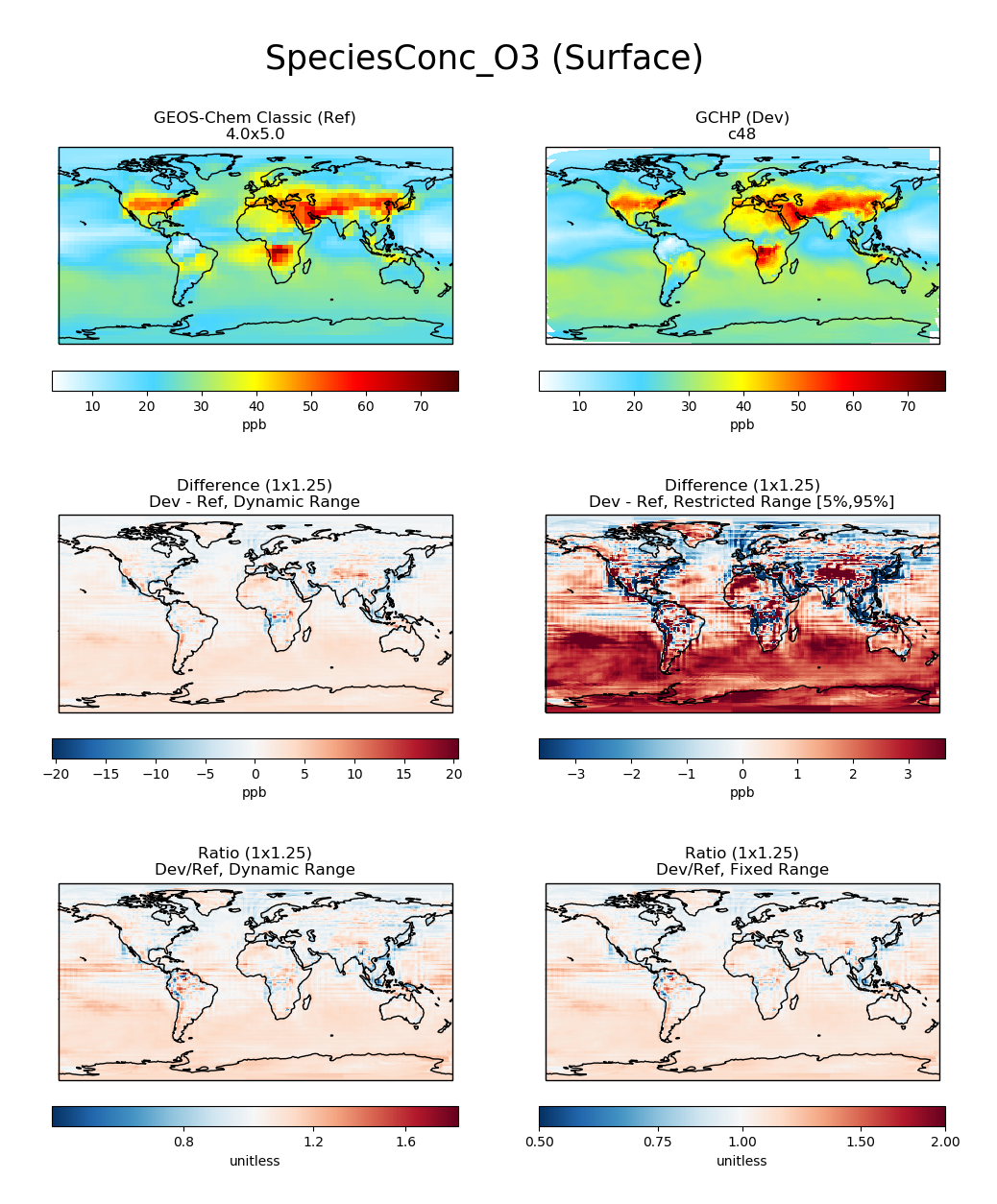
Function compare_zonal_mean¶
def compare_zonal_mean(
refdata,
refstr,
devdata,
devstr,
varlist=None,
itime=0,
refmet=None,
devmet=None,
weightsdir='.',
pdfname="",
cmpres=None,
match_cbar=True,
pres_range=None,
normalize_by_area=False,
enforce_units=True,
convert_to_ugm3=False,
flip_ref=False,
flip_dev=False,
use_cmap_RdBu=False,
verbose=False,
log_color_scale=False,
log_yaxis=False,
extra_title_txt=None,
n_job=-1,
sigdiff_list=None,
second_ref=None,
second_dev=None,
spcdb_dir=os.path.dirname(__file__),
sg_ref_path='',
sg_dev_path='',
ref_vert_params=None,
dev_vert_params=None,
**extra_plot_args
):
"""
Creates 3x2 comparison zonal-mean plots for variables
common in two xarray Datasets. Optionally save to PDF.
Args:
refdata: xarray dataset
Dataset used as reference in comparison
refstr: str
String description for reference data to be used in plots
devdata: xarray dataset
Dataset used as development in comparison
devstr: str
String description for development data to be used in plots
Keyword Args (optional):
varlist: list of strings
List of xarray dataset variable names to make plots for
Default value: None (will compare all common 3D variables)
itime: integer
Dataset time dimension index using 0-based system
Default value: 0
refmet: xarray dataset
Dataset containing ref meteorology
Default value: None
devmet: xarray dataset
Dataset containing dev meteorology
Default value: None
weightsdir: str
Directory path for storing regridding weights
Default value: None (will create/store weights in
current directory)
pdfname: str
File path to save plots as PDF
Default value: Empty string (will not create PDF)
cmpres: str
String description of grid resolution at which
to compare datasets
Default value: None (will compare at highest resolution
of Ref and Dev)
match_cbar: bool
Set this flag to True to use same the colorbar bounds
for both Ref and Dev plots.
Default value: True
pres_range: list of two integers
Pressure range of levels to plot [hPa]. The vertical axis
will span the outer pressure edges of levels that contain
pres_range endpoints.
Default value: [0, 2000]
normalize_by_area: bool
Set this flag to True to to normalize raw data in both
Ref and Dev datasets by grid area. Input ref and dev
datasets must include AREA variable in m2 if normalizing
by area.
Default value: False
enforce_units: bool
Set this flag to True force an error if the variables in
the Ref and Dev datasets have different units.
Default value: True
convert_to_ugm3: str
Whether to convert data units to ug/m3 for plotting.
Default value: False
flip_ref: bool
Set this flag to True to flip the vertical dimension of
3D variables in the Ref dataset.
Default value: False
flip_dev: bool
Set this flag to True to flip the vertical dimension of
3D variables in the Dev dataset.
Default value: False
use_cmap_RdBu: bool
Set this flag to True to use a blue-white-red colormap for
plotting raw reference and development datasets.
Default value: False
verbose: logical
Set this flag to True to enable informative printout.
Default value: False
log_color_scale: bool
Set this flag to True to enable plotting data (not diffs)
on a log color scale.
Default value: False
log_yaxis: bool
Set this flag to True if you wish to create zonal mean
plots with a log-pressure Y-axis.
Default value: False
extra_title_txt: str
Specifies extra text (e.g. a date string such as "Jan2016")
for the top-of-plot title.
Default value: None
n_job: int
Defines the number of simultaneous workers for parallel
plotting. Set to 1 to disable parallel plotting.
Value of -1 allows the application to decide.
Default value: -1
sigdiff_list: list of str
Returns a list of all quantities having significant
differences (where |max(fractional difference)| > 0.1).
Default value: None
second_ref: xarray Dataset
A dataset of the same model type / grid as refdata,
to be used in diff-of-diffs plotting.
Default value: None
second_dev: xarray Dataset
A dataset of the same model type / grid as devdata,
to be used in diff-of-diffs plotting.
Default value: None
spcdb_dir: str
Directory containing species_database.yml file.
Default value: Path of GCPy code repository
sg_ref_path: str
Path to NetCDF file containing stretched-grid info
(in attributes) for the ref dataset
Default value: '' (will not be read in)
sg_dev_path: str
Path to NetCDF file containing stretched-grid info
(in attributes) for the dev dataset
Default value: '' (will not be read in)
ref_vert_params: list(AP, BP) of list-like types
Hybrid grid parameter A in hPa and B (unitless).
Needed if ref grid is not 47 or 72 levels.
Default value: None
dev_vert_params: list(AP, BP) of list-like types
Hybrid grid parameter A in hPa and B (unitless).
Needed if dev grid is not 47 or 72 levels.
Default value: None
extra_plot_args: various
Any extra keyword arguments are passed through the
plotting functions to be used in calls to pcolormesh()
(CS) or imshow() (Lat/Lon).
"""
and generates a comparison plot such as:
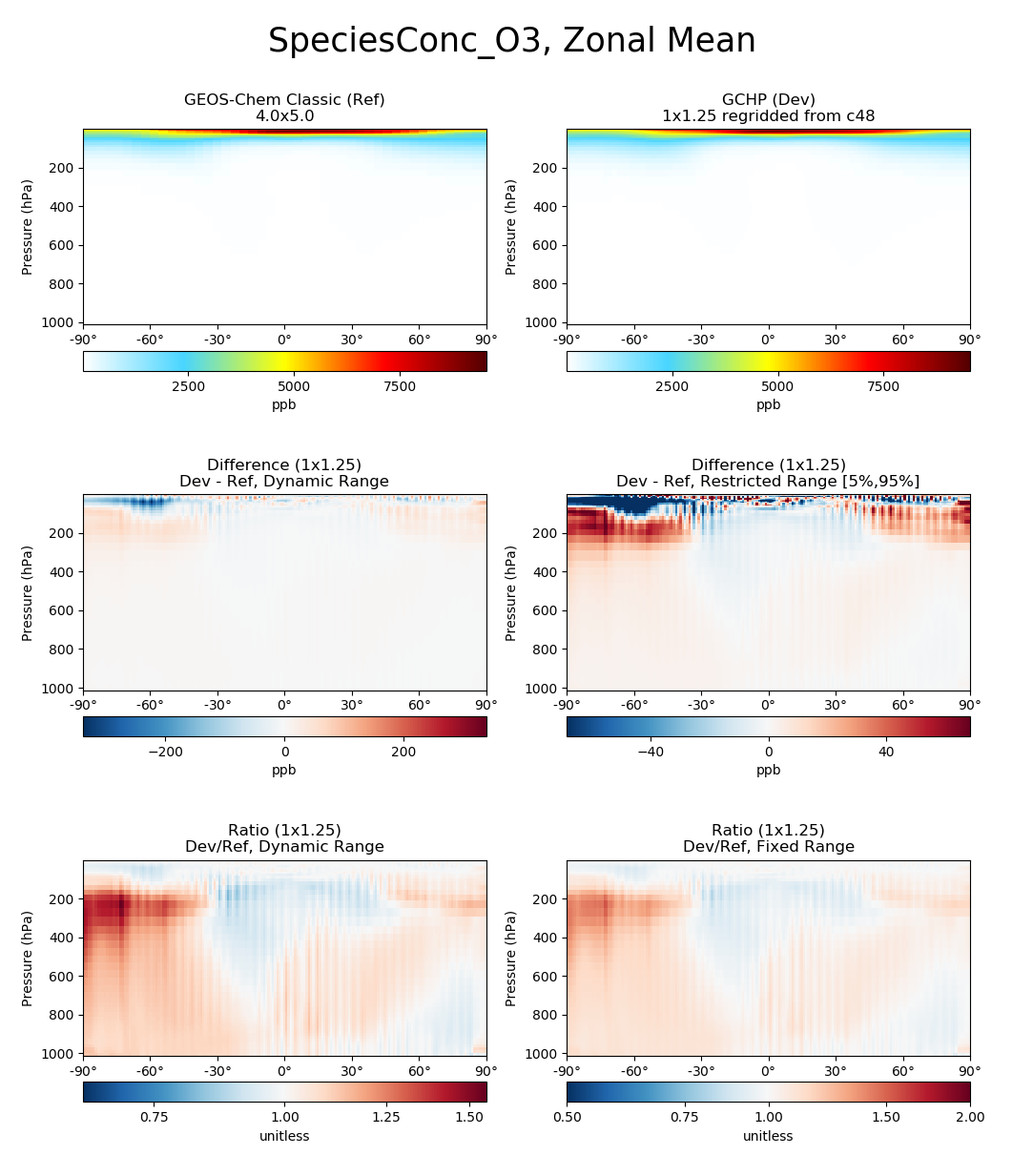
Example script¶
Here is a basic script that calls both compare_zonal_mean() and
compare_single_level():
#!/usr/bin/env python
import xarray as xr
import matplotlib.pyplot as plt
from gcpy.plot.compare_single_level import compare_single_level
from gcpy.plot.compare_zonal_mean import compare_zonal_mean
file1 = '/path/to/ref'
file2 = '/path/to/dev'
ds1 = xr.open_dataset(file1)
ds2 = xr.open_dataset(file2)
compare_zonal_mean(ds1, 'Ref run', ds2, 'Dev run')
plt.show()
compare_single_level(ds1, 'Ref run', ds2, 'Dev run')
plt.show()
Single panel plots¶
Function single_panel() (contained in GCPy module
gcpy.plot.single_panel) is used to create plots containing
only one panel of GEOS-Chem data. This function is used within
compare_single_level() and compare_zonal_mean() to
generate each panel plot. It can also be called directly on its
own to quickly plot GEOS-Chem data in zonal mean or single level
format.
Function: single_panel¶
Function single_panel() accepts the following arguments:
def single_panel(
plot_vals,
ax=None,
plot_type="single_level",
grid=None,
gridtype="",
title="fill",
comap=WhGrYlRd,
norm=None,
unit="",
extent=None,
masked_data=None,
use_cmap_RdBu=False,
log_color_scale=False,
add_cb=True,
pres_range=None,
pedge=np.full((1, 1), -1),
pedge_ind=np.full((1, 1), -1),
log_yaxis=False,
xtick_positions=None,
xticklabels=None,
proj=ccrs.PlateCarree(),
sg_path='',
ll_plot_func="imshow",
vert_params=None,
pdfname="",
weightsdir='.',
vmin=None,
vmax=None,
return_list_of_plots=False,
**extra_plot_args
):
"""
Core plotting routine -- creates a single plot panel.
Args:
plot_vals: xarray.DataArray, numpy.ndarray, or dask.array.Array
Single data variable GEOS-Chem output to plot
Keyword Args (Optional):
ax: matplotlib axes
Axes object to plot information
Default value: None (Will create a new axes)
plot_type: str
Either "single_level" or "zonal_mean"
Default value: "single_level"
grid: dict
Dictionary mapping plot_vals to plottable coordinates
Default value: {} (will attempt to read grid from plot_vals)
gridtype: str
"ll" for lat/lon or "cs" for cubed-sphere
Default value: "" (will automatically determine from grid)
title: str
Title to put at top of plot
Default value: "fill" (will use name attribute of plot_vals
if available)
comap: matplotlib Colormap
Colormap for plotting data values
Default value: WhGrYlRd
norm: list
List with range [0..1] normalizing color range for matplotlib
methods. Default value: None (will determine from plot_vals)
unit: str
Units of plotted data
Default value: "" (will use units attribute of plot_vals
if available)
extent: tuple (minlon, maxlon, minlat, maxlat)
Describes minimum and maximum latitude and longitude of input
data. Default value: None (Will use full extent of plot_vals
if plot is single level).
masked_data: numpy array
Masked area for avoiding near-dateline cubed-sphere plotting
issues Default value: None (will attempt to determine from
plot_vals)
use_cmap_RdBu: bool
Set this flag to True to use a blue-white-red colormap
Default value: False
log_color_scale: bool
Set this flag to True to use a log-scale colormap
Default value: False
add_cb: bool
Set this flag to True to add a colorbar to the plot
Default value: True
pres_range: list(int)
Range from minimum to maximum pressure for zonal mean
plotting. Default value: [0, 2000] (will plot entire
atmosphere)
pedge: numpy array
Edge pressures of vertical grid cells in plot_vals
for zonal mean plotting. Default value: np.full((1, 1), -1)
(will determine automatically)
pedge_ind: numpy array
Index of edge pressure values within pressure range in
plot_vals for zonal mean plotting.
Default value: np.full((1, 1), -1) (will determine
automatically)
log_yaxis: bool
Set this flag to True to enable log scaling of pressure in
zonal mean plots. Default value: False
xtick_positions: list(float)
Locations of lat/lon or lon ticks on plot
Default value: None (will place automatically for
zonal mean plots)
xticklabels: list(str)
Labels for lat/lon ticks
Default value: None (will determine automatically from
xtick_positions)
proj: cartopy projection
Projection for plotting data
Default value: ccrs.PlateCarree()
sg_path: str
Path to NetCDF file containing stretched-grid info
(in attributes) for plot_vals.
Default value: '' (will not be read in)
ll_plot_func: str
Function to use for lat/lon single level plotting with
possible values 'imshow' and 'pcolormesh'. imshow is much
faster but is slightly displaced when plotting from dateline
to dateline and/or pole to pole. Default value: 'imshow'
vert_params: list(AP, BP) of list-like types
Hybrid grid parameter A in hPa and B (unitless). Needed if
grid is not 47 or 72 levels. Default value: None
pdfname: str
File path to save plots as PDF
Default value: "" (will not create PDF)
weightsdir: str
Directory path for storing regridding weights
Default value: "." (will store regridding files in
current directory)
vmin: float
minimum for colorbars
Default value: None (will use plot value minimum)
vmax: float
maximum for colorbars
Default value: None (will use plot value maximum)
return_list_of_plots: bool
Return plots as a list. This is helpful if you are using
a cubedsphere grid and would like access to all 6 plots
Default value: False
extra_plot_args: various
Any extra keyword arguments are passed to calls to
pcolormesh() (CS) or imshow() (Lat/Lon).
Returns:
plot: matplotlib plot
Plot object created from input
"""
Function single_panel() expects data with a 1-length (or
non-existent) T (time) dimension, as well as a
1-length or non-existent Z (vertical level) dimension.
single_panel() contains a few amenities to help with plotting
GEOS-Chem data, including automatic grid detection for lat/lon or
standard cubed-sphere xarray DataArray-s. You can also pass NumPy
arrays to plot, though you’ll need to manually pass grid info in this
case (with the gridtype, pedge, and
pedge_ind keyword arguments).
The sample script shown below shows how you can data at a single level and
timestep from an xarray.DataArray object.
#!/usr/bin/env python
import xarray as xr
import matplotlib.pyplot as plt
from gcpy.plot.single_panel import single_panel
# Read data from a file into an xr.Dataset object
dset = xr.open_dataset('GEOSChem.SpeciesConc.20160701_0000z.nc4')
# Extract ozone (v/v) from the xr.Dataset object,
# for time=0 (aka first timestep) and lev=0 (aka surface)
sfc_o3 = dset['SpeciesConcVV_O3'].isel(time=0).isel(lev=0)
# Plot the data!
single_panel(sfc_o3)
plt.show()